AI-generated Key Takeaways
-
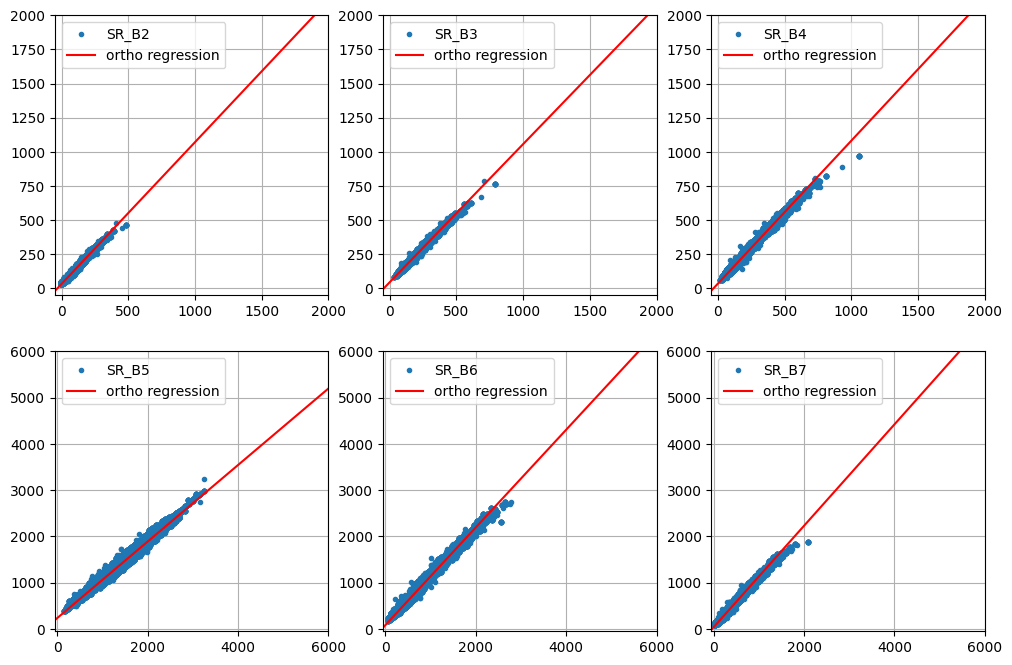
Relative radiometric normalization is essential for comparing pixel intensities across different image acquisitions and time series analysis, especially when images are uncalibrated or poorly corrected.
-
The tutorial uses orthogonal linear regression on "no-change" pixels, identified by the iMAD algorithm, to determine the radiometric relationship between two images.
-
The regression coefficients derived from the orthogonal regression are applied to normalize one image (target) to another (reference).
-
Relative radiometric normalization, referred to as harmonization in the context of different missions, can be applied to data from different satellite sensors like Landsat 8 and Landsat 9.
-
The presented examples demonstrate the application of this method for normalizing Sentinel-2 TOA to SR images and harmonizing Landsat 8 and Landsat 9 surface reflectance data.
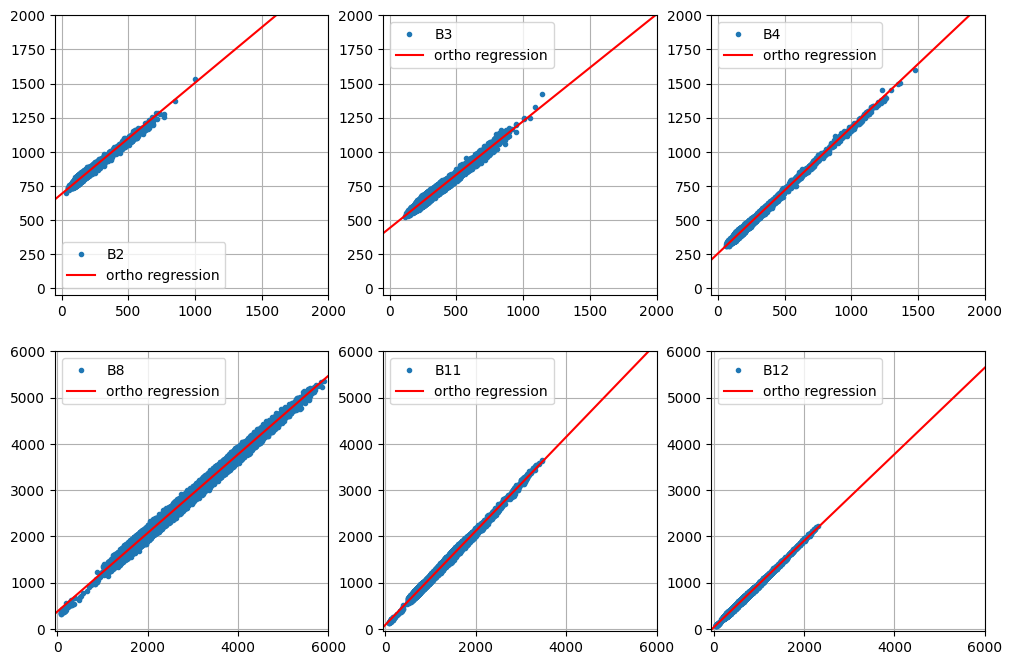
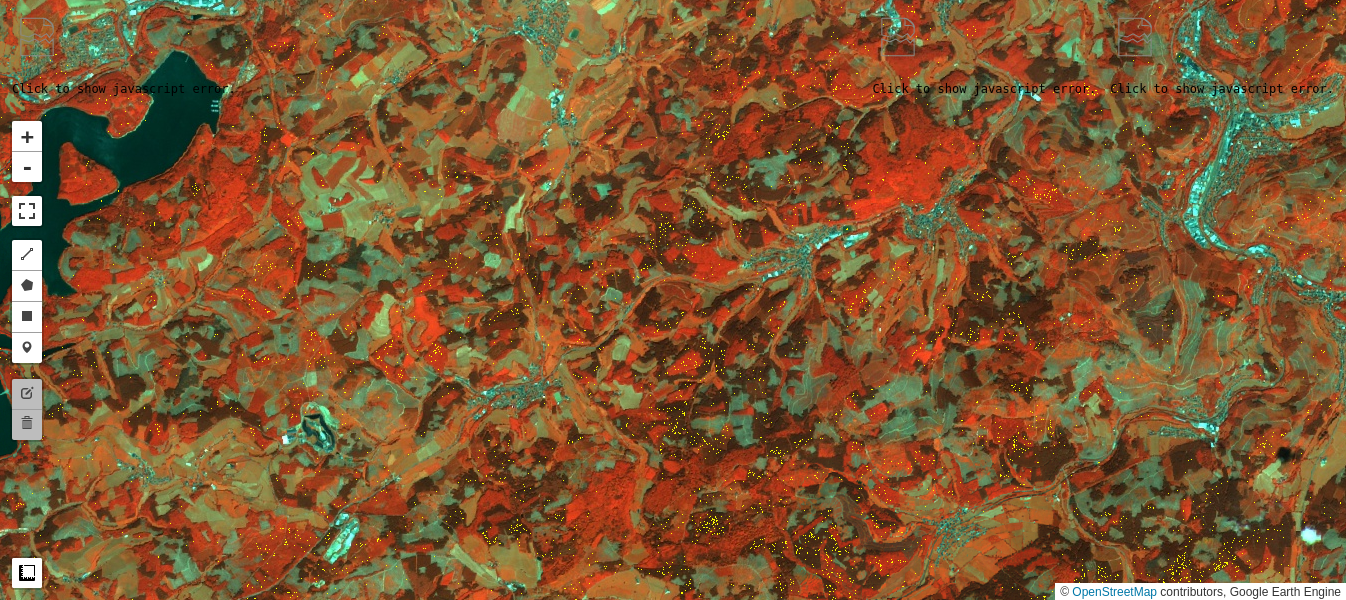
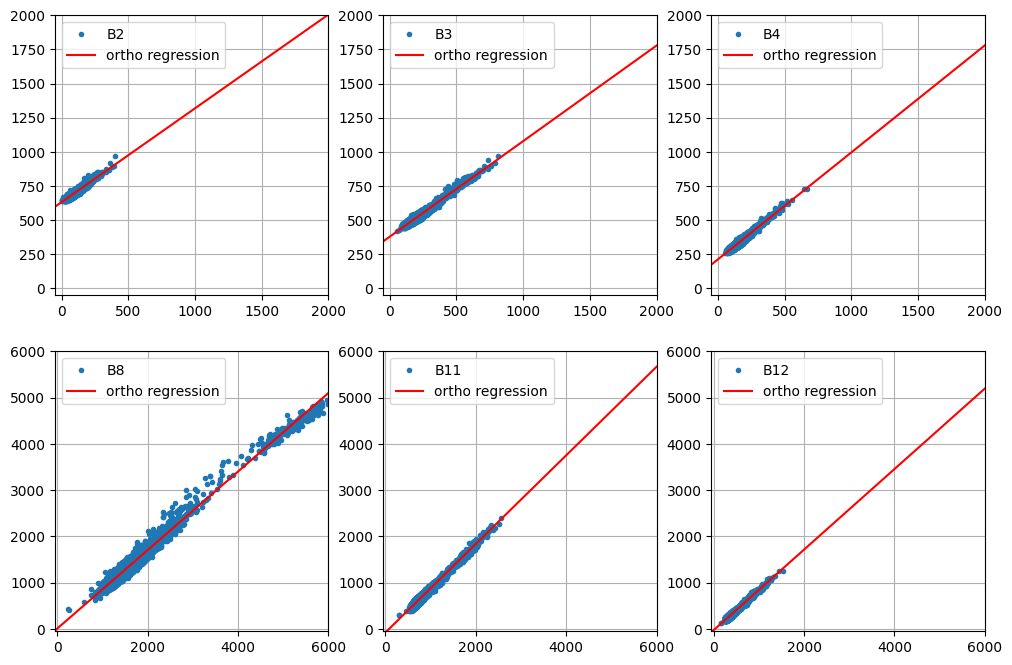
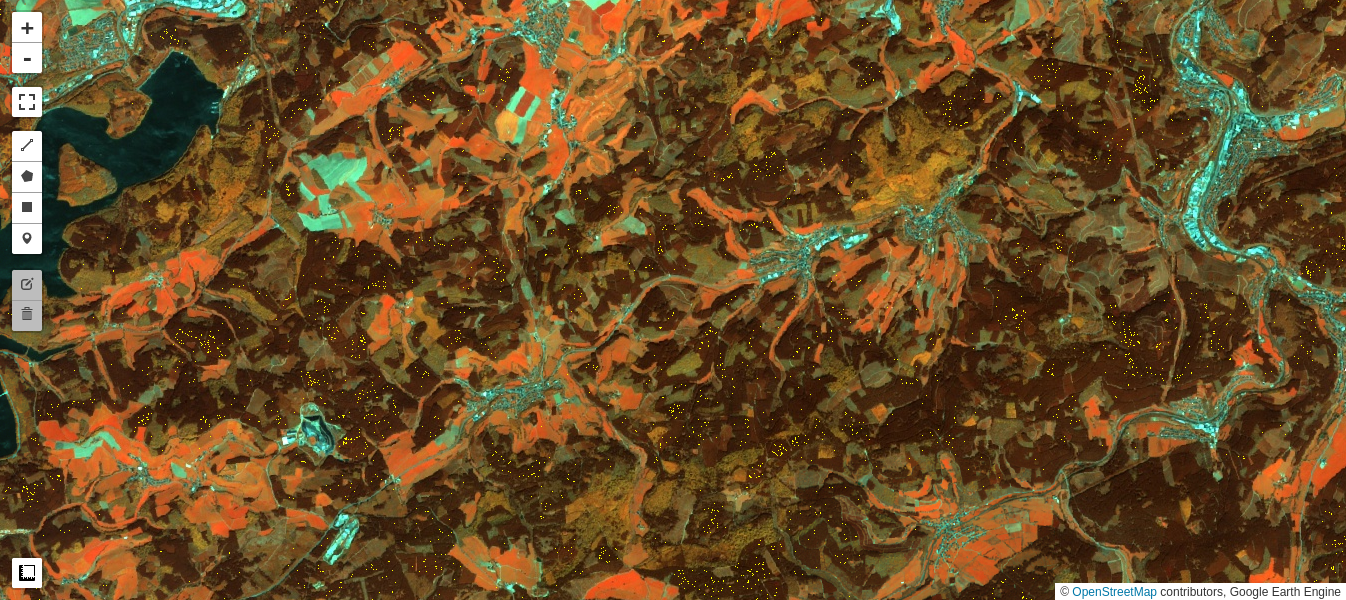
Test of pixel intensities across different image acquisitions requires that the received signals have similar radiometric scales. As we saw in Part 2 of the present tutorial, provided the iMAD algorithm converges satisfactorily, it will isolate the pixels which have not significantly changed in the two input images presented to it. A regression analysis on these no-change pixels can then determine how well the radiometric parameters of the two acquisitions compare. If, for instance, images that were corrected to absolute surface reflectance are examined in this way, we would expect a good match, i.e., a slope close to one and an intercept close to zero at all spectral wavelengths. In general, for uncalibrated or poorly corrected images, this will not be the case. Then, the regression coefficients can be applied to normalize one image (the target, say) to the other (the reference). This is a prerequisite, for example, for tracing features such as NDVI indices over a time series when the images have not been reduced to surface reflectances, see e.g., Gan et al. (2021) , or indeed for harmonizing the data from two different sensors of the same family, such as Landsat 8 with Landsat 9. In this final part of the GEE Change Detection Tutorial we'll have a closer look at this approach, which we'll refer to here as relative radiometric normalization . For more detailed treatment, see also Canty and Nielsen (2008) , Canty et al. (2004) , and Schroeder et al. (2006) . A different (but similar) method involving identification of so-called pseudoinvariant features is explained in another GEE community tutorial in this series.
Preliminaries
This tutorial will optionally export assets. Edit the following EXPORT_PATH
variable to specify the location to store the assets.
All assets that are needed to complete the tutorial are hosted by Earth Engine,
but if you'd like to display assets that you export, replace paths as needed.
# Enter your own export to assets path name here -----------
EXPORT_PATH
=
'projects/YOUR_GEE_PROJECT_NAME/assets/imad/'
# ------------------------------------------------
import
ee
ee
.
Authenticate
(
auth_mode
=
'notebook'
)
ee
.
Initialize
()
# Import other packages used in the tutorial
%
matplotlib
inline
import
geemap
import
numpy
as
np
import
random
,
time
import
matplotlib.pyplot
as
plt
from
scipy.stats
import
norm
,
chi2
from
pprint
import
pprint
# for pretty printing
Routines from Parts 1 and 2
def
trunc
(
values
,
dec
=
3
):
'''Truncate a 1-D array to dec decimal places.'''
return
np
.
trunc
(
values
*
10
**
dec
)
/
(
10
**
dec
)
# Display an image in a one percent linear stretch.
def
display_ls
(
image
,
map
,
name
,
centered
=
False
):
bns
=
image
.
bandNames
()
.
length
()
.
getInfo
()
if
bns
==
3
:
image
=
image
.
rename
(
'B1'
,
'B2'
,
'B3'
)
pb_99
=
[
'B1_p99'
,
'B2_p99'
,
'B3_p99'
]
pb_1
=
[
'B1_p1'
,
'B2_p1'
,
'B3_p1'
]
img
=
ee
.
Image
.
rgb
(
image
.
select
(
'B1'
),
image
.
select
(
'B2'
),
image
.
select
(
'B3'
))
else
:
image
=
image
.
rename
(
'B1'
)
pb_99
=
[
'B1_p99'
]
pb_1
=
[
'B1_p1'
]
img
=
image
.
select
(
'B1'
)
percentiles
=
image
.
reduceRegion
(
ee
.
Reducer
.
percentile
([
1
,
99
]),
maxPixels
=
1e11
)
mx
=
percentiles
.
values
(
pb_99
)
if
centered
:
mn
=
ee
.
Array
(
mx
)
.
multiply
(
-
1
)
.
toList
()
else
:
mn
=
percentiles
.
values
(
pb_1
)
map
.
addLayer
(
img
,
{
'min'
:
mn
,
'max'
:
mx
},
name
)
def
covarw
(
image
,
weights
=
None
,
scale
=
20
,
maxPixels
=
1e10
):
'''Return the centered image and its weighted covariance matrix.'''
try
:
geometry
=
image
.
geometry
()
bandNames
=
image
.
bandNames
()
N
=
bandNames
.
length
()
if
weights
is
None
:
weights
=
image
.
constant
(
1
)
weightsImage
=
image
.
multiply
(
ee
.
Image
.
constant
(
0
))
.
add
(
weights
)
means
=
image
.
addBands
(
weightsImage
)
\ .
reduceRegion
(
ee
.
Reducer
.
mean
()
.
repeat
(
N
)
.
splitWeights
(),
scale
=
scale
,
maxPixels
=
maxPixels
)
\ .
toArray
()
\ .
project
([
1
])
centered
=
image
.
toArray
()
.
subtract
(
means
)
B1
=
centered
.
bandNames
()
.
get
(
0
)
b1
=
weights
.
bandNames
()
.
get
(
0
)
nPixels
=
ee
.
Number
(
centered
.
reduceRegion
(
ee
.
Reducer
.
count
(),
scale
=
scale
,
maxPixels
=
maxPixels
)
.
get
(
B1
))
sumWeights
=
ee
.
Number
(
weights
.
reduceRegion
(
ee
.
Reducer
.
sum
(),
geometry
=
geometry
,
scale
=
scale
,
maxPixels
=
maxPixels
)
.
get
(
b1
))
covw
=
centered
.
multiply
(
weights
.
sqrt
())
\ .
toArray
()
\ .
reduceRegion
(
ee
.
Reducer
.
centeredCovariance
(),
geometry
=
geometry
,
scale
=
scale
,
maxPixels
=
maxPixels
)
\ .
get
(
'array'
)
covw
=
ee
.
Array
(
covw
)
.
multiply
(
nPixels
)
.
divide
(
sumWeights
)
return
(
centered
.
arrayFlatten
([
bandNames
]),
covw
)
except
Exception
as
e
:
print
(
'Error:
%s
'
%
e
)
def
corr
(
cov
):
'''Transform covariance matrix to correlation matrix.'''
# Diagonal matrix of inverse sigmas.
sInv
=
cov
.
matrixDiagonal
()
.
sqrt
()
.
matrixToDiag
()
.
matrixInverse
()
# Transform.
corr
=
sInv
.
matrixMultiply
(
cov
)
.
matrixMultiply
(
sInv
)
.
getInfo
()
# Truncate.
return
[
list
(
map
(
trunc
,
corr
[
i
]))
for
i
in
range
(
len
(
corr
))]
def
geneiv
(
C
,
B
):
'''Return the eigenvalues and eigenvectors of the generalized eigenproblem
C*X = lambda*B*X'''
try
:
C
=
ee
.
Array
(
C
)
B
=
ee
.
Array
(
B
)
# Li = choldc(B)^-1
Li
=
ee
.
Array
(
B
.
matrixCholeskyDecomposition
()
.
get
(
'L'
))
.
matrixInverse
()
# Solve symmetric, ordinary eigenproblem Li*C*Li^T*x = lambda*x
Xa
=
Li
.
matrixMultiply
(
C
)
\ .
matrixMultiply
(
Li
.
matrixTranspose
())
\ .
eigen
()
# Eigenvalues as a row vector.
lambdas
=
Xa
.
slice
(
1
,
0
,
1
)
.
matrixTranspose
()
# Eigenvectors as columns.
X
=
Xa
.
slice
(
1
,
1
)
.
matrixTranspose
()
# Generalized eigenvectors as columns, Li^T*X
eigenvecs
=
Li
.
matrixTranspose
()
.
matrixMultiply
(
X
)
return
(
lambdas
,
eigenvecs
)
except
Exception
as
e
:
print
(
'Error:
%s
'
%
e
)
# Collect a Sentinel-2 image pair.
def
collect
(
aoi
,
t1a
,
t1b
,
t2a
,
t2b
,
coll1
,
coll2
=
None
):
try
:
if
coll2
==
None
:
coll2
=
coll1
im1
=
ee
.
Image
(
ee
.
ImageCollection
(
coll1
)
.
filterBounds
(
aoi
)
.
filterDate
(
ee
.
Date
(
t1a
),
ee
.
Date
(
t1b
))
.
filter
(
ee
.
Filter
.
contains
(
rightValue
=
aoi
,
leftField
=
'.geo'
))
.
sort
(
'CLOUDY_PIXEL_PERCENTAGE'
)
.
sort
(
'CLOUD_COVER_LAND'
)
.
first
()
.
clip
(
aoi
)
)
im2
=
ee
.
Image
(
ee
.
ImageCollection
(
coll2
)
.
filterBounds
(
aoi
)
.
filterDate
(
ee
.
Date
(
t2a
),
ee
.
Date
(
t2b
))
.
filter
(
ee
.
Filter
.
contains
(
rightValue
=
aoi
,
leftField
=
'.geo'
))
.
sort
(
'CLOUDY_PIXEL_PERCENTAGE'
)
.
sort
(
'CLOUD_COVER_LAND'
)
.
first
()
.
clip
(
aoi
)
)
timestamp
=
im1
.
date
()
.
format
(
'E MMM dd HH:mm:ss YYYY'
)
print
(
timestamp
.
getInfo
())
timestamp
=
im2
.
date
()
.
format
(
'E MMM dd HH:mm:ss YYYY'
)
print
(
timestamp
.
getInfo
())
return
(
im1
,
im2
)
except
Exception
as
e
:
print
(
'Error:
%s
'
%
e
)
def
chi2cdf
(
Z
,
df
):
'''Chi-square cumulative distribution function with df degrees of freedom.'''
return
ee
.
Image
(
Z
.
divide
(
2
))
.
gammainc
(
ee
.
Number
(
df
)
.
divide
(
2
))
def
imad
(
current
,
prev
):
'''Iterator function for iMAD.'''
done
=
ee
.
Number
(
ee
.
Dictionary
(
prev
)
.
get
(
'done'
))
return
ee
.
Algorithms
.
If
(
done
,
prev
,
imad1
(
current
,
prev
))
def
imad1
(
current
,
prev
):
'''Iteratively re-weighted MAD.'''
image
=
ee
.
Image
(
ee
.
Dictionary
(
prev
)
.
get
(
'image'
))
Z
=
ee
.
Image
(
ee
.
Dictionary
(
prev
)
.
get
(
'Z'
))
allrhos
=
ee
.
List
(
ee
.
Dictionary
(
prev
)
.
get
(
'allrhos'
))
nBands
=
image
.
bandNames
()
.
length
()
.
divide
(
2
)
weights
=
chi2cdf
(
Z
,
nBands
)
.
subtract
(
1
)
.
multiply
(
-
1
)
scale
=
ee
.
Dictionary
(
prev
)
.
getNumber
(
'scale'
)
niter
=
ee
.
Dictionary
(
prev
)
.
getNumber
(
'niter'
)
# Weighted stacked image and weighted covariance matrix.
centeredImage
,
covarArray
=
covarw
(
image
,
weights
,
scale
)
bNames
=
centeredImage
.
bandNames
()
bNames1
=
bNames
.
slice
(
0
,
nBands
)
bNames2
=
bNames
.
slice
(
nBands
)
centeredImage1
=
centeredImage
.
select
(
bNames1
)
centeredImage2
=
centeredImage
.
select
(
bNames2
)
s11
=
covarArray
.
slice
(
0
,
0
,
nBands
)
.
slice
(
1
,
0
,
nBands
)
s22
=
covarArray
.
slice
(
0
,
nBands
)
.
slice
(
1
,
nBands
)
s12
=
covarArray
.
slice
(
0
,
0
,
nBands
)
.
slice
(
1
,
nBands
)
s21
=
covarArray
.
slice
(
0
,
nBands
)
.
slice
(
1
,
0
,
nBands
)
c1
=
s12
.
matrixMultiply
(
s22
.
matrixInverse
())
.
matrixMultiply
(
s21
)
b1
=
s11
c2
=
s21
.
matrixMultiply
(
s11
.
matrixInverse
())
.
matrixMultiply
(
s12
)
b2
=
s22
# Solution of generalized eigenproblems.
lambdas
,
A
=
geneiv
(
c1
,
b1
)
_
,
B
=
geneiv
(
c2
,
b2
)
rhos
=
lambdas
.
sqrt
()
.
project
(
ee
.
List
([
1
]))
# Test for convergence.
lastrhos
=
ee
.
Array
(
allrhos
.
get
(
-
1
))
done
=
rhos
.
subtract
(
lastrhos
)
\ .
abs
()
\ .
reduce
(
ee
.
Reducer
.
max
(),
ee
.
List
([
0
]))
\ .
lt
(
ee
.
Number
(
0.0001
))
\ .
toList
()
\ .
get
(
0
)
allrhos
=
allrhos
.
cat
([
rhos
.
toList
()])
# MAD variances.
sigma2s
=
rhos
.
subtract
(
1
)
.
multiply
(
-
2
)
.
toList
()
sigma2s
=
ee
.
Image
.
constant
(
sigma2s
)
# Ensure sum of positive correlations between X and U is positive.
tmp
=
s11
.
matrixDiagonal
()
.
sqrt
()
ones
=
tmp
.
multiply
(
0
)
.
add
(
1
)
tmp
=
ones
.
divide
(
tmp
)
.
matrixToDiag
()
s
=
tmp
.
matrixMultiply
(
s11
)
.
matrixMultiply
(
A
)
.
reduce
(
ee
.
Reducer
.
sum
(),
[
0
])
.
transpose
()
A
=
A
.
matrixMultiply
(
s
.
divide
(
s
.
abs
())
.
matrixToDiag
())
# Ensure positive correlation.
tmp
=
A
.
transpose
()
.
matrixMultiply
(
s12
)
.
matrixMultiply
(
B
)
.
matrixDiagonal
()
tmp
=
tmp
.
divide
(
tmp
.
abs
())
.
matrixToDiag
()
B
=
B
.
matrixMultiply
(
tmp
)
# Canonical and MAD variates.
centeredImage1Array
=
centeredImage1
.
toArray
()
.
toArray
(
1
)
centeredImage2Array
=
centeredImage2
.
toArray
()
.
toArray
(
1
)
U
=
ee
.
Image
(
A
.
transpose
())
.
matrixMultiply
(
centeredImage1Array
)
\ .
arrayProject
([
0
])
\ .
arrayFlatten
([
bNames1
])
V
=
ee
.
Image
(
B
.
transpose
())
.
matrixMultiply
(
centeredImage2Array
)
\ .
arrayProject
([
0
])
\ .
arrayFlatten
([
bNames2
])
iMAD
=
U
.
subtract
(
V
)
# Chi-square image.
Z
=
iMAD
.
pow
(
2
)
\ .
divide
(
sigma2s
)
\ .
reduce
(
ee
.
Reducer
.
sum
())
return
ee
.
Dictionary
({
'done'
:
done
,
'scale'
:
scale
,
'niter'
:
niter
.
add
(
1
),
'image'
:
image
,
'allrhos'
:
allrhos
,
'Z'
:
Z
,
'iMAD'
:
iMAD
})
def
run_imad
(
aoi
,
image1
,
image2
,
assetId
,
scale
=
10
,
maxiter
=
100
):
try
:
N
=
image1
.
bandNames
()
.
length
()
.
getInfo
()
imadnames
=
[
'iMAD'
+
str
(
i
+
1
)
for
i
in
range
(
N
)]
imadnames
.
append
(
'Z'
)
# Maximum iterations.
inputlist
=
ee
.
List
.
sequence
(
1
,
maxiter
)
first
=
ee
.
Dictionary
({
'done'
:
0
,
'scale'
:
scale
,
'niter'
:
ee
.
Number
(
0
),
'image'
:
image1
.
addBands
(
image2
),
'allrhos'
:
[
ee
.
List
.
sequence
(
1
,
N
)],
'Z'
:
ee
.
Image
.
constant
(
0
),
'iMAD'
:
ee
.
Image
.
constant
(
0
)})
# Iteration.
result
=
ee
.
Dictionary
(
inputlist
.
iterate
(
imad
,
first
))
# Retrieve results.
iMAD
=
ee
.
Image
(
result
.
get
(
'iMAD'
))
.
clip
(
aoi
)
rhos
=
ee
.
String
.
encodeJSON
(
ee
.
List
(
result
.
get
(
'allrhos'
))
.
get
(
-
1
))
Z
=
ee
.
Image
(
result
.
get
(
'Z'
))
niter
=
result
.
getNumber
(
'niter'
)
# Export iMAD and Z as a singe image, including rhos and number of iterations in properties.
iMAD_export
=
ee
.
Image
.
cat
(
iMAD
,
Z
)
.
rename
(
imadnames
)
.
set
(
'rhos'
,
rhos
,
'niter'
,
niter
)
assexport
=
ee
.
batch
.
Export
.
image
.
toAsset
(
iMAD_export
,
description
=
'assetExportTask'
,
assetId
=
assetId
,
scale
=
scale
,
maxPixels
=
1e10
)
assexport
.
start
()
print
(
'Exporting iMAD to
%s
\n
task id:
%s
'
%
(
assetId
,
str
(
assexport
.
id
)))
except
Exception
as
e
:
print
(
'Error:
%s
'
%
e
)